Life on Film by Stephen Fortune
Thu 03 Oct 2013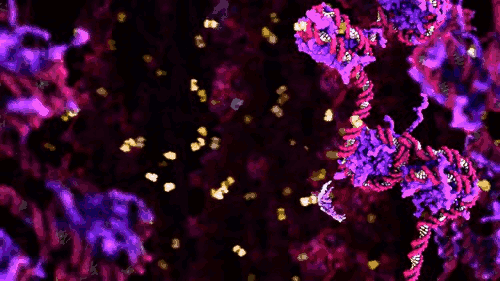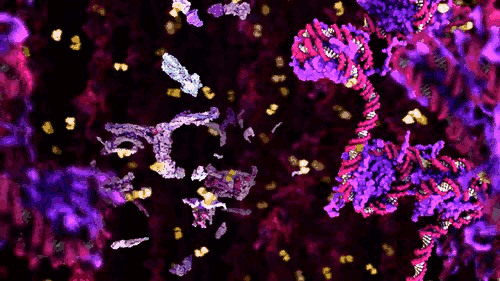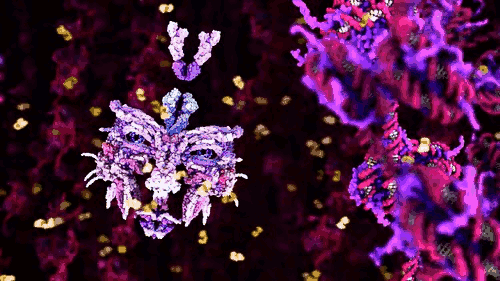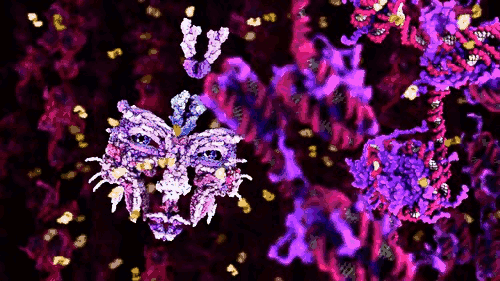
Stephen Fortune has kindly written us an insightful essay with accompanying gifs, meditating and expanding upon Life on Film, an intriguing collection complied specially for AND, which will be screened in the AND Hub on Fri 04 Oct.
Each piece within ‘Life on Film’ unveils biological processes. Each work concerns itself with revealing one or more of the inapprehensible processes of life. As in most forms of documentary none of the assembled pieces contend to be an exhaustive testimony of the totality of ‘life’ and living things. Since the innovation of Robert Hooke’s microscope [1] the historical trajectory of the biological sciences has been inseparable from an intensifying awareness of the lively and living processes that elude visible apprehension.
‘Life on Film’s chronological progression from microcinematography to present day data visualisation is a timeline of exponentially sophisticated mediating technologies [2]. This essay would like to prompt consideration of how each life-mediating technology operates as epistemic objects (objects, artifacts or tools integral to the construction of empirical proof, and subsequently knowledge). To paraphrase Werner Heisenberg,
“We have to remember that what we observe is not life herself, but life exposed to our method of questioning.”
This program chronicles the life science’s paradigm of ‘drilling down’ into progressively deeper layers of living mechanisms; from cells in motion, to organelles and “molecular machines”, to DNA and its methylated metadata (aka epigenetics). The trajectory of technological tools applied to biology has followed one of “aletheia”. The Greek word Plato deployed in his famous cave allegory can be translated as “uncovering” or “the state of not being hidden” (rather than the conventional translation of “truth”). It’s an important translational distinction for appreciating the function of representational apparatuses within the life sciences. The pieces bookending the programme illustrate this.
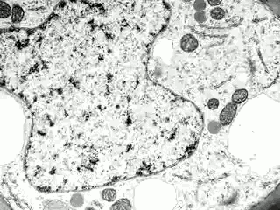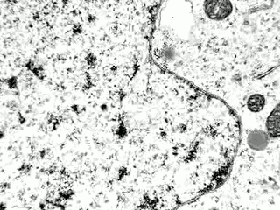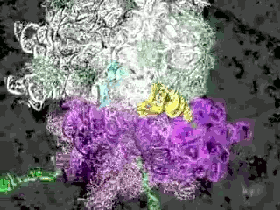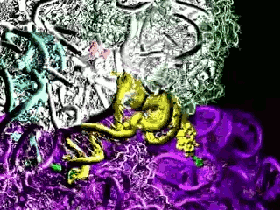
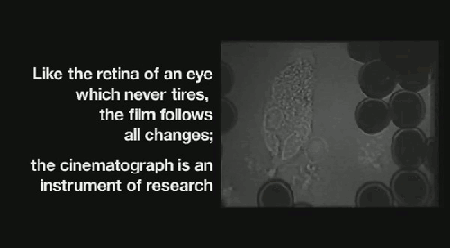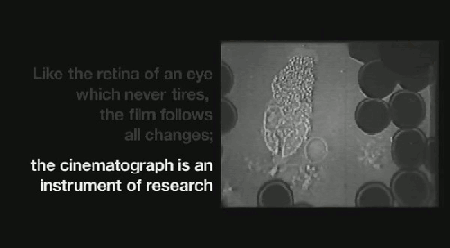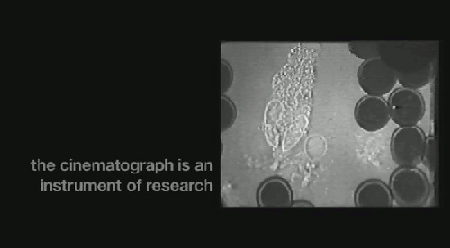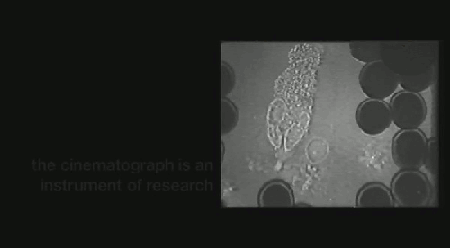 ‘Life on Film’ begins (aptly) with “News About Cells” (pictured above), Hannah Landecker & Chris Kelty’s compilation of early 20th century time-lapse science footage, known then as microcinematography. Microcinematography laid bare the movement of cell cultures, over spanned duration, for the first time. One attribute of living matter was isolated, observed and provided basis for deeper scientific comprehension.
‘Life on Film’ begins (aptly) with “News About Cells” (pictured above), Hannah Landecker & Chris Kelty’s compilation of early 20th century time-lapse science footage, known then as microcinematography. Microcinematography laid bare the movement of cell cultures, over spanned duration, for the first time. One attribute of living matter was isolated, observed and provided basis for deeper scientific comprehension.
The programme concludes with Stanford’s Whole Cell data visualisation (pictured below) of the humble STD ‘mycoplasma genitalium’. Here over 900 papers worth of empirical data, derived using multiple and varied experimental apparatuses, have been aggregated into an interactive edifice which extrapolates multiple informational dimensions about a living organism. Gene expression, life cycles, and metabolism all play out as dynamic data visualisations, unveiling facets of life through thoroughly in-silico remediation (it is possible for 11,336 total visualisations of life through this technology). The gaze of data mining uncovers life procedures that evade visible detection.
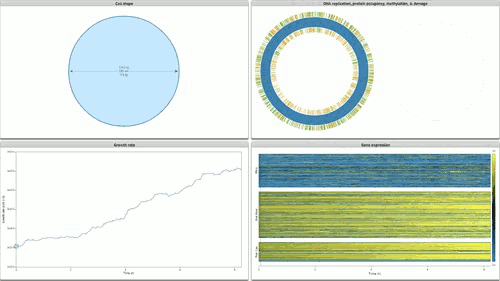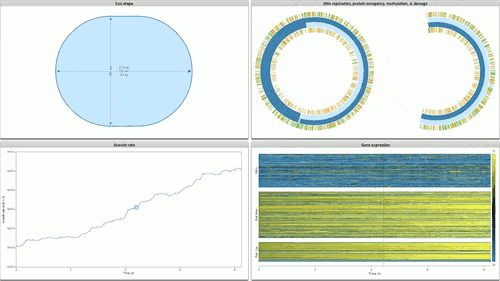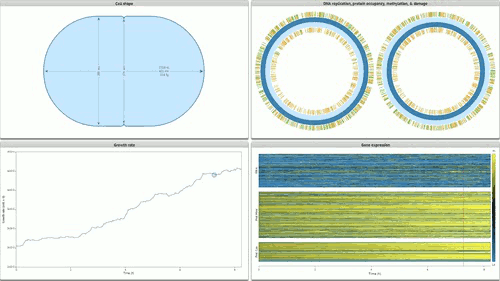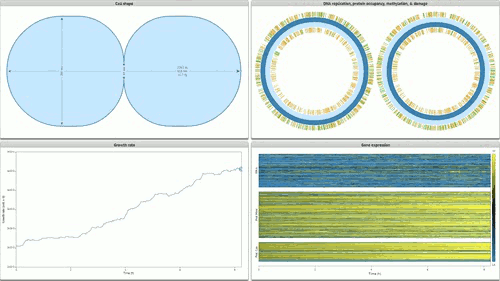 Time-lapse cinema is wide reaching in the cycles of life it illuminates: everything from larvae maturation, to the unfurling of plants in response to the sun are unveiled by sampling and compressing the duration of life documented. Hannah Landecker’s research in the domain of living cell microcinematography demonstrates that sampling organisms at periodic intervals is integral to comprehending heretofore unseen mechanisms of life. The term sampling is preferred here because frequency sampling allows us to understand the direct link between an established cultural form (cinema) and how digital technologies parse and remediate the processes and substrates they are capable of parsing as ‘media’.
Time-lapse cinema is wide reaching in the cycles of life it illuminates: everything from larvae maturation, to the unfurling of plants in response to the sun are unveiled by sampling and compressing the duration of life documented. Hannah Landecker’s research in the domain of living cell microcinematography demonstrates that sampling organisms at periodic intervals is integral to comprehending heretofore unseen mechanisms of life. The term sampling is preferred here because frequency sampling allows us to understand the direct link between an established cultural form (cinema) and how digital technologies parse and remediate the processes and substrates they are capable of parsing as ‘media’.
Appreciating the mechanisms of how digital technologies produce an object for reflection, interpretation or education is key to apprehending the intersection of digital (aka ‘in-silico’) logics and biological logics which typifies the contemporary condition of the molecular life sciences, and its well publicised sub-discipline of synthetic biology. How frequently an analog input, be it an MP3 record, cell life-cycles or protein folding, is made discreet is essential to each of the remediations of life displayed on screen.
 Another of the supercomputer simulations makes this explicit. The Ribosome simulations of the Sanbonmatsu Laboratory (pictured above) are explicitly intended to simulate the motions of a ribosome by building an atom by atom modelling of the ribosome at work. Here an incredibly short duration, 10 nanoseconds, is slowed down and expanded to aid human comprehension. The ribosome is a crucial part of our cells – it is the cellular complex which converts the nucleic acid information contained in DNA sequences into functional proteins comprised of amino acids.
Another of the supercomputer simulations makes this explicit. The Ribosome simulations of the Sanbonmatsu Laboratory (pictured above) are explicitly intended to simulate the motions of a ribosome by building an atom by atom modelling of the ribosome at work. Here an incredibly short duration, 10 nanoseconds, is slowed down and expanded to aid human comprehension. The ribosome is a crucial part of our cells – it is the cellular complex which converts the nucleic acid information contained in DNA sequences into functional proteins comprised of amino acids.
These simulations, dating from 2005, are nevertheless instructive in terms of the digital infrastructure requisite to their realisation: 768 of the 8,192 available processors of the Los Alamos National Laboratory’s supercomputer ‘Q” machine’ were required to simulate 10 nanoseconds of ribosome activity! The expenditure of digital computation necessary to reveal the operations of the ‘biological analogue of a CPU’ merits contemplation in and of itself.
 Together with the Cytochrome Rollercoaster and the Stanford Whole Cell Visualisation these three pieces represent a (very) potted history of the increasing use of computation to unveil the processes integral to life over the last three decades. The inclusion of the KLUGE visualisation system points (pictured above) to the earliest intersection of intracellular macromolecular entities (e.g. proteins) and graphics simulation software.
Together with the Cytochrome Rollercoaster and the Stanford Whole Cell Visualisation these three pieces represent a (very) potted history of the increasing use of computation to unveil the processes integral to life over the last three decades. The inclusion of the KLUGE visualisation system points (pictured above) to the earliest intersection of intracellular macromolecular entities (e.g. proteins) and graphics simulation software.
Eric Francouer, who researched the system informs us that this was the first practical use of such graphics systems within scientific practice. The ball and stick models provided a way of representing the three dimensional form of a protein. Proteins are the elements of cells that allow it to function and exist – everything living depends on protein function. The structure of a protein is integral to its utility as a molecular machine. Said structure is predominantly deduced through the technology of X-Ray crystallography (2013 marks the centenary of this scientific method).
With X-Ray crystallography successive snapshots of a crystallised protein accrete into a static 3D model. The model toured by the ‘cytochrome rollercoaster’ is similarly static, despite the novel choice of manifesting the 3D structure of the protein by bringing the viewer on a visceral first-person journey through a cytochrome proteins bends and folds. Though newer technologies exist x-ray crystallography is one of the most ubiquitous means of representing the intracellular realm.
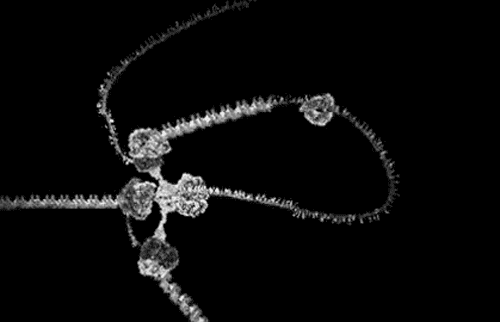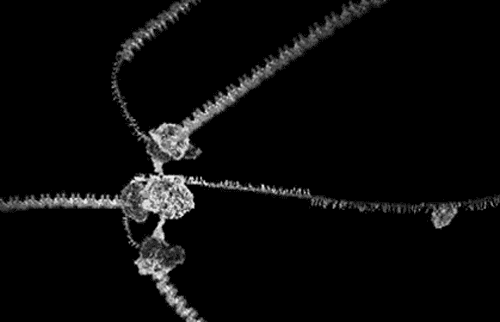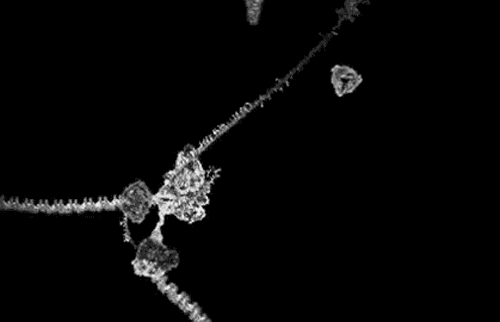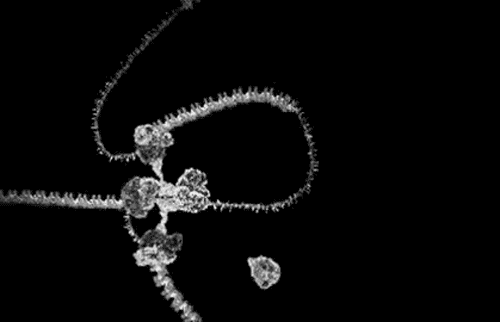 The award winning computer animations of Drew Berry (pictured above) draw their scientific veracity from just that method. It is a mode of biological comprehension which has attracted critical consideration from the field of science & technology studies [3]. At it’s simplest, intracellular information is omitted by the process: “what cannot be crystallized does not exist as an epistemic object for this technology”[4]. At the more nuanced and problematic is the issue that the models produced, as visible in the KLUGE system, render a dynamic, constantly in motion entity, as static (albeit malleable per a computer interface).
The award winning computer animations of Drew Berry (pictured above) draw their scientific veracity from just that method. It is a mode of biological comprehension which has attracted critical consideration from the field of science & technology studies [3]. At it’s simplest, intracellular information is omitted by the process: “what cannot be crystallized does not exist as an epistemic object for this technology”[4]. At the more nuanced and problematic is the issue that the models produced, as visible in the KLUGE system, render a dynamic, constantly in motion entity, as static (albeit malleable per a computer interface).
This exact caveat is noted by Nobel Laureate Paul Berg in his introduction to “An Epic on the Cellular Level”. The solution which this video posits is as novel as it is entertaining: a trippy performative dance reenactment of the process of cellular transcription – the aforementioned ‘computation’ which the ribosome enacts upon DNA ‘information’. The chaotic ebbs and flows of human bodies in motion arguably articulates the messy dynamism of intracellular existence as authentically as Berry’s award winning animations.
The acknowledgement of models as ‘necessarily limited’ by Berg, coupled with rendering a cell through human bodies, rather than simulated atoms, is a pointed reminder that our models for apprehending the fundamental operations of life are just that – models. Natasha Myers is among the forefront of critical thinkers addressing the epistemic consequences of rendering such dynamic entities as static. The correlationism between computation and molecular machines which distinguishes most of the synthetic biology discourse from prior paradigms would do well to heed the lessons therein.
 Synthetic biology features within this programme through the protocell circus corralled by Rachel Armstrong & Michael Simon Toon. Armstrong’s protocells (pictured above) [5] are not living, they are ‘pure chemistry’ that apes some of the features recognised as ‘life-like’. What is displayed underscores another problematic inherent to representing and comprehending life, as Rheingberger articulates:
Synthetic biology features within this programme through the protocell circus corralled by Rachel Armstrong & Michael Simon Toon. Armstrong’s protocells (pictured above) [5] are not living, they are ‘pure chemistry’ that apes some of the features recognised as ‘life-like’. What is displayed underscores another problematic inherent to representing and comprehending life, as Rheingberger articulates:
“What is being measured. Is it still a biological function, or has it shrunk to a chemical process?”
Intersections : Some thoughts on instruments and objects in the experimental context of the life sciences, Rheinberger, H.-J.
Michael Joaquin Grey’s hypnotic video art piece “Artificial muscle contraction (misconception)” [6] recalls a related paradigm shifting moment in the biological sciences, by exhibiting the self assembly behaviour of muscle proteins ex-vivo. In-vivo and in-vitro (or ex-vivo) are biological designations which refer to whether the cell is within the organism, or abstracted outside it (and have inspired the ‘in-silico’ designation for software that works with biological matter). This piece evokes a landmark experiment (Eduard Buchner’s efforts to obtain a cell-free alcoholic fermentation enzyme) which introduced those designations to the biological sciences. As Rheinberger notes:
“From now on biologists distinguished between in vitro and in vivo experiments. With the in vitro experiment a vitreous envelope is created which replaces the … wraps of the organism.What Claude Bernard termed the “milieu intérieur,” the inner environment of the organic processes, is replaced by a chemo-technical milieu that at the same time opens a new analytical space; a space turned inside out, tipped over, and trimmed for new connections.”
Intersections : Some thoughts on instruments and objects in the experimental context of the life sciences, Rheinberger, H.-J.
It is hoped that the collected works of Life on Film extend Rheinberger’s (above) problematic into the mediation of life. For in addition to documenting the processes of life, the dynamic visual content on screen are produced by tools which instigate and imply an epistemic reframing of life. ‘Seeing’ begets comprehension, from which knowledge for manipulation and reconstruction of the subject of enquiry can be derived. For “in-silico” simulation, optimisation, and aggregation is integral not only comprehending life, but increasingly it is involved in synthesising it as well [7].
 The visual chronicling of life proceeds at the cutting edge of technologies of seeing, and beyond in some cases: the Whole Cell visualisation ushers in a totally new register of seeing, the gaze afforded by data mining, and biological “aletheia” continues to add more tools to its repertoire, each successive tool aspiring to involve less and less overt ‘touch’ of the represented subject.
The visual chronicling of life proceeds at the cutting edge of technologies of seeing, and beyond in some cases: the Whole Cell visualisation ushers in a totally new register of seeing, the gaze afforded by data mining, and biological “aletheia” continues to add more tools to its repertoire, each successive tool aspiring to involve less and less overt ‘touch’ of the represented subject.
In there lies a whole other facet of the cinema of biology: the link between sophistication of light control technologies and both the deeper comprehension (as is possible through confocal microscopy and manipulation) of living things. For now, and in the case of all the ‘Life on Film’ content, the represented subject in question must be ‘touched’ in some manner to be rendered perceivable. There follows poetic consequences in addition to the already noted epistemic ramifications. To pick two instances: practicing scientists are aware that their software systems exhibit simulated pressures not present in real world molecular dynamics.
As theorists like Emily Candela indicate, artists can extrapolate this to a hedonics of touch vis a vis the imperceptible realm of molecular biology. Or the ever sophisticating tools can, as Verena Friedrich work illustrates, permit artists to venture into media-mimetic biology, where living matter exists as a substrate possessed of unique animating affordances.
Footnotes:
1.
As this masterful Philip Ball essay elaborates, the effect of those early forays was ontological shock (and therefore we might consider ontological PTSD) for culture and thinkers in general. It is curious indeed that a comparable ontological shock for our society had to be staged by collectives such as CAE and Tissue Culture and Art (TC&A) Collective
2.
The issues of classical aesthetics (which collecting these works together in a screening programme does invoke) are sidestepped for the most part – a collection of essays (collated by guest editor Daisy Ginsberg) in Current Opinion in Chemical Biology spans the breadth of many of the issues touched upon in this essay, with deeper consideration of classical aesthetics theory, and I would recommend it as essential additional literature for the curious reader.
3. This is but one of many recommended starting points – click here.
4.
Intersections : Some thoughts on instruments and objects in the experimental context of the life sciences. Rheinberger, H.-J.
5.
The science of protocells goes all the way back to Stephane Leduc, who aimed to use them to illustrate that life was a fundamentally chemical process. As recent bioartwork ‘Mechanism of Life’ by Catts, Zurr & van Sice [http://www.isea2013.org/events/semipermeable-plus/] illustrates, Leduc first coined the term ‘synthetic biology’ to denote his methodology. The ancestry of the synthetic biology epistemic concept has largely been ignored by contemporary engineers in the field, an issue which ‘Mechanism of Life’ goes some way towards addressing.
6.
(traditionally displayed as a loop)
7.
The same bundle of wires, internet cabling, logic gates and user interfaces which make Whole Cell Visualisation a knowledge resource equally enables novel scientific entities like eteRNA. eteRNA is a game where players make combinations of RNA – a problem that is hard for computers but easy for humans. Successful ‘high score’ combinations are synthesised Stanford biochemistry lab as an additional step in verifying their utility as wetware.
Recent Journals
- A Gig at Sunrise: Reflecting on W Brzask at Ephemera Festival
- Announcing our THREE FIELDS artists
- New Rhythms
- Introducing Commons // Keiken and Jazmin Morris
- Introducing our Creative Associates programme
- Reflections on the Associate Board Member Programme
- The Future of Arts Governance
- Rendering our virtual, net and digital discourses
- Announcing a new partnership between AND and the School of Digital Arts
Other Journals
-
2025
-
2024
-
2023
-
2022
-
2021
-
2020
-
2019
-
2018
-
2017
-
2016
-
2015
-
2014
-
2013
-
2012
-
2011



